Cellular-level Alterations in Epileptogenesis - MR Microscopy of Organotypic Hippocampal Slice Cultures
- 1. University Medical Center Freiburg, Dept. of Radiology, Medical Physics, Freiburg im Breisgau, Germany
- 2. University of Freiburg, BrainLinks-BrainTools, Cluster of Excellence, Freiburg im Breisgau, Germany
- 3. University Medical Center Freiburg, Experimental Epilepsy Research, Freiburg im Breisgau, Germany
- 4. University of Freiburg, Dept. of Microsystems Engineering - IMTEK, Freiburg im Breisgau, Germany
- 5. Karlsruhe Institute of Technology, Institute of Microstructure Technology IMT, Karlsruhe, Germany
Organotypic hippocampal slice cultures (OHSC) are a well established neuronal culture system that combines the advantages of cell culturing with a neuronal network tightly reflecting the in vivo state [1]. We aim to investigate structural changes during epileptogenesis in OHSC by using high spatial resolution MR microscopy, which allows continuous monitoring near/at the cellular level [2] [3]. Here we demonstrate high-resolution structural imaging of fixed hippocampal slices as a first step to resolve hippocampal cytoarchitecture and connectivity patterns of epileptic and healthy tissue.
A commercially available mousehead two-element quadrature cryogenic coil system with a 7 T Bruker BioSpec 70/20 small animal scanner was used to adapt MR pulse sequences with respect to OHSC imaging. Up to now, seven fixed slices (400 µm thick) of kainate injected (epileptic) and control animals were then inserted into a PMMA container filled with saline solution and sealed on both sides with adhesive PCR tape to avoid evaporation. Multi-slice 2D gradient echo sequences were applied with: TR = 300 ms, TE= 13 ms, flip angle = 50°, resolution 18×18×104 µm³ obtained in 2h 56min. An adapted spin-echo EPI DTI sequence was used for DTI measurements with TR = 3000 ms, TE= 39 ms, resolution 39×39×100 µm³, NEX = 16, 60 directions, scan time = 3h 31min. The neuronal cytoarchitecture was subsequently compared to fluorescence microscopy of immunohistochemically stained tissue sections.
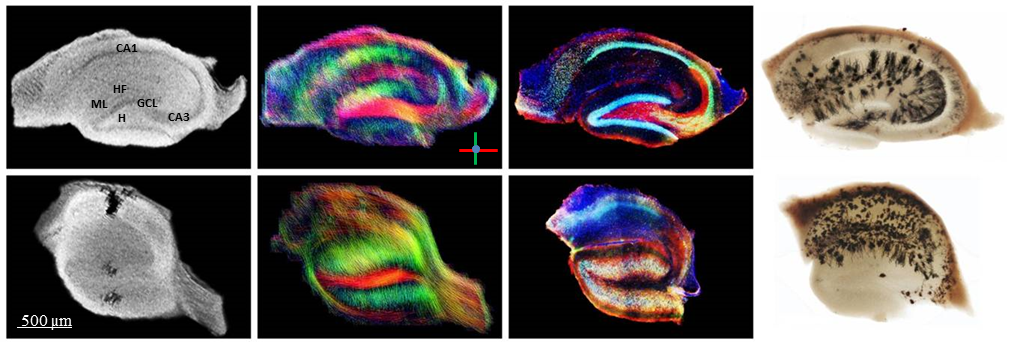
The morphological MR images reveal strong differences between the epileptic and the healthy control slices: While in the control slice the densely packed neuronal cell layers (cornu ammonis: CA, granule cell layer: GCL) can easily be identified, the GCL of the epileptic animal is strongly dispersed. The strongest and most consistent DTI signals can be found in areas of parallel oriented dendritic trees (of granule cells and pyramidal cells) as well as in areas of axonal fiber pathways. The connectivity pattern within the slice of the epileptic animal is strongly changed; the dendritic tree signals of CA1 pyramidal cells seem to be almost completely lost, which is confirmed by the MRI and immunohistochemical data.
A fundamental understanding of these processes is a necessity to overcome technological challenges associated with in vivo studies as well as to find new markers for an early diagnosis and the specific treatment of epilepsy.
Funding: DFG grant EXC 1086.
- [1] X. Chai, G. Münzer et al., (2014), Epilepsy-induced motility of differentiated neurons, Cereb Cortex, 2130-40, 24(8)
- [2] J.J. Flint et al., (2012), Magnetic resonance microscopy of human and porcine neurons and cellular processes, NeuroImage, 1404-1411, 60
- [3] K. Göbel et al., (2014), MR Microscopy and DTI of Organotypic Hippocampal Slice Cultures, In: Proc ISMRM, Milan, Italy
- [4] N. Gogolla, (2006), Preparation of organotypic hippocampal slice cultures for long-term live imaging, Nature Protocols, 1165-1171, 1
